Welcome to Iris Biotech
For better service please confirm your country and language we detected.

For better service please confirm your country and language we detected.

Thank you very much for your interest in our products. All prices listed on our website are ex-works, Germany, and may attract customs duties when imported.
You may/will be contacted by the shipping company for additional documentation that may be required by the US Customs for clearance.
We offer you the convenience of buying through a local partner, Peptide Solutions LLC who can import the shipment as well as prepay the customs duties and brokerage on your behalf and provide the convenience of a domestic sale.
Continue to Iris Biotech GmbHSend request to US distributorChemischer Name: N-(4-hydroxyphenethyl)-6-((4R,5S)-5-methyl-2-oxoimidazolidin-4-yl)hexanamide // Synonyme: Desthiobiotin Phenol, (4R,5S)-oxo-N-[2-(4-hydroxyphenyl)ethyl]-5-methyl-2-4-Imidazolidinehexanamide
Startet von 375,00 €
Biotin-Tyramide analog that binds less tightly to biotin-binding proteins. It can be displaced competitively by biotin. Streptavidin-based ligands can be gently stripped from desthiobiotin-tyramide-labeled targets with buffered biotin solutions.
Mapping the Proteome of the Synaptic Cleft through Proximity Labeling Reveals New Cleft Proteins; T. Cijsouw, A. Ramsey, T. Lam, B. Carbone, T. Blanpied and T. Biederer; 2018; 6: 48. https://www.mdpi.com/2227-7382/6/4/48
Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation; V. Hung, S. S. Lam, N. D. Udeshi, T. Svinkina, G. Guzman, V. K. Mootha, S. A. Carr and A. Y. Ting; Elife D. Pagliarini 2017; 6: e24463. https://doi.org/10.7554/eLife.24463
In Situ Peroxidase Labeling and Mass-Spectrometry Connects Alpha-Synuclein Directly to Endocytic Trafficking and mRNA Metabolism in Neurons; C. Y. Chung, V. Khurana, S. Yi, N. Sahni, K. H. Loh, P. K. Auluck, V. Baru, N. D. Udeshi, Y. Freyzon, S. A. Carr, D. E. Hill, M. Vidal, A. Y. Ting and S. Lindquist; Cell Syst 2017; 4: 242-250 e4. https://doi.org/10.1016/j.cels.2017.01.002
Identification of Microprotein-Protein Interactions via APEX Tagging; Q. Chu, A. Rathore, J. K. Diedrich, C. J. Donaldson, J. R. Yates, 3rd and A. Saghatelian; Biochemistry 2017; 56: 3299-3306. https://doi.org/10.1021/acs.biochem.7b00265
Proximity-dependent labeling methods for proteomic profiling in living cells; C. L. Chen and N. Perrimon; Wiley Interdiscip Rev Dev Biol 2017; 6: e272. https://doi.org/10.1002/wdev.272
Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts; K. H. Loh, P. S. Stawski, A. S. Draycott, N. D. Udeshi, E. K. Lehrman, D. K. Wilton, T. Svinkina, T. J. Deerinck, M. H. Ellisman, B. Stevens, S. A. Carr and A. Y. Ting; Cell 2016; 166: 1295-1307 e21. https://doi.org/10.1016/j.cell.2016.07.041
Directed evolution of APEX2 for electron microscopy and proximity labeling; S. S. Lam, J. D. Martell, K. J. Kamer, T. J. Deerinck, M. H. Ellisman, V. K. Mootha and A. Y. Ting; Nat Methods 2015; 12: 51-4. https://doi.org/10.1038/nmeth.3179
New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay; X. W. Li, J. S. Rees, P. Xue, H. Zhang, S. W. Hamaia, B. Sanderson, P. E. Funk, R. W. Farndale, K. S. Lilley, S. Perrett and A. P. Jackson; J Biol Chem 2014; 289: 14434-47. https://doi.org/10.1074/jbc.M113.529578
Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging; H. W. Rhee, P. Zou, N. D. Udeshi, J. D. Martell, V. K. Mootha, S. A. Carr and A. Y. Ting; Science 2013; 339: 1328-1331. https://doi.org/10.1126/science.1230593
Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry; M. R. Clutter, G. C. Heffner, P. O. Krutzik, K. L. Sachen and G. P. Nolan; Cytometry A 2010; 77: 1020-31. https://doi.org/10.1002/cyto.a.20970
A. J. Gross and I. W. Sizer; J. Biol. Chem. 1959; 234: 1611.
Sie benötigen größere Mengen für Ihre Entwicklung oder Produktion?
Bitte senden Sie mir mehr Informationen über

Herein, we present 2-Iminobiotin, a cyclic guanidino analogue of biotin with pH-dependent affinity for avidin, and compare it to biotin and desthio
Details ansehen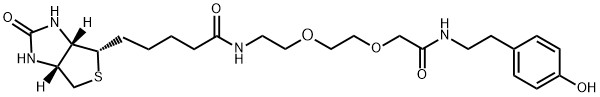
Methods for proteomic mapping of cellular organelles or subdomains that avoid the necessity of cell lysis and purification by targeting specific su
Details ansehen






