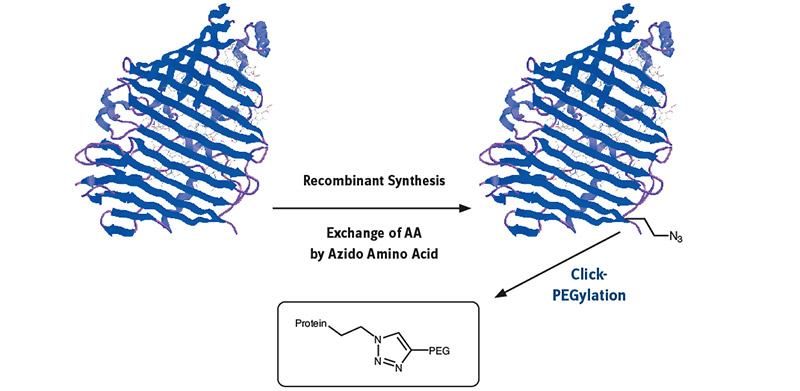
Mutant or modified aminoacyl tRNA synthetases (aaRS) have been used to charge non-natural amino acids to the corresponding tRNA, which incorporates them into polypeptides or proteins during recombinant synthesis.
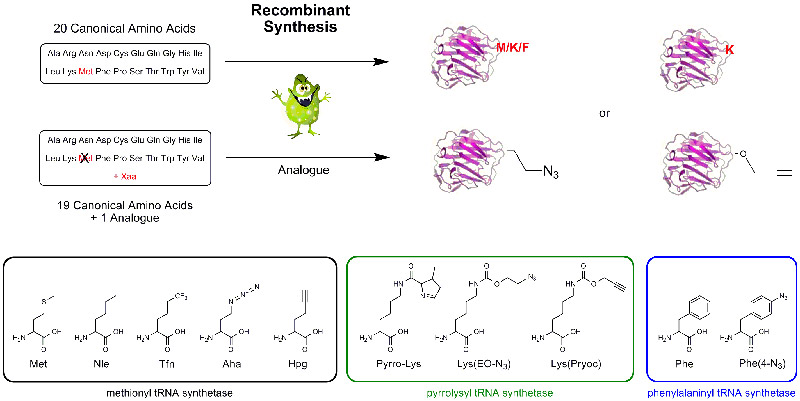
a) Recombinant Incorporation of Amino Acids into Proteins
Mutant or modified aminoacyl tRNA synthetases (aaRS) have been used to charge non-natural amino acids to the corresponding tRNA, which incorporates them into polypeptides or proteins during recombinant synthesis. As azido homoalanine (Aha) is a structure analogue of methionine (Met), Met has been replaced effectively by Aha in proteins by the native methionyl tRNA synthetase of E. coli. A modified yeast phenylalanyl-tRNA synthetase was prepared by introduction of a T415G or T415A mutation in its α-subunit; specific residues in tryptophanyl-tRNA synthetase and methionyl-tRNA synthetase are also identified, where mutagenesis results in specificity for non-natural aminoacyl-tRNA molecules. tRNA can be loaded with azido and propargyl analogues of lysine by pyrrolysyl-tRNA synthetase enabling the biochemist to introduce site specifically an azido or alkyne group into a protein for further click conjugation. Further developments and inventions have been done to specifically incorporate a vast number of Click components into practically any protein.
Once the azido function has been built into the protein sequence, conjugation with a large number of diverse partners opens a wide field of possibilities. Many different applications from therapeutics to diagnostics can be addressed through conjugates with PEG-polymers, dyes, cofactors, antibodies, small molecules, toxins, additional proteins, and peptides.
References:
- Incorporation of Azides into Rrecombinant Proteins for Chemoselective Modification by the Staudinger ligation; Kristi L. Kiick, Eliana Saxon, David A. Tirrell, and Carolyn R. Bertozzi; Proc. Natl. Acad. Sci. USA 2002; 99: 19.
- Patent: WO 2007103307
- High-Throughput Screening for Methionyl-tRNA Synthetases that Enable Residue-Specific Incorporation of Noncanonical Amino Acids into Recombinant Proteins in Bacterial Cells; Tae Hyeon Yoo and David A. Tirrell; Angew. Chem. Int. Ed. 2007; 46: 5340–5343; DOI: 10.1002/anie.200700779.
- Global Replacement of Tryptophan with Aminotryptophans Generates Non-Invasive Protein-Based Optical pH Sensors; Nediljko Budisa, Marina Rubini, Jae H. Bae, Elisabeth Weyher, Waltraud Wenger, Ralph Golbik, Robert Huber, Luis Moroder; Angew. Chem. Int. Ed. 2002; 41(21): 4066-4069.
- Genetic Encoding and Labeling of Aliphatic Azides and Alkynes in Recombinant Proteins via a Pyrrolysyl-tRNA Synthetase/tRNA(CUA) Pair and Click Chemistry; Duy P. Nguyen, Hrvoje Lusic, Heinz Neumann, Prashant B. Kapadnis, Alexander Deiters, and Jason W. Chin; J. Am. Chem. Soc. 2009; 131: 8720-8721.
- Direct charging of tRNACUA with pyrrolysine in vitro and in vivo; Blight Sherry et. al; Nature 2004; 431: 333-335.
Protocol for Protein Labelling by Cycloaddition:
- Dilute the desired proteome sample to a 2 mg/ml solution in PBS.
- Add 500 µl of diluted 2 mg/ml proteome solution to an Eppendorf tube.
- Add 1–20 µM of the desired alkyne probe from a concentrated DMSO stock: do not exceed 10 µl of DMSO in this 500 µl reaction. After addition of the probe, vortex and leave at room temperature for 1 h.
- Prepare a fresh solution of 50 µM TCEP (LS-3405) in water (14.4 mg/ml) and a 1.7 mM stock of ligand in DMSO:tBuOH 1:4 (0.9 mg/ml). Store TCEP at 4°C under argon. Stock solution in water should always be prepared freshly.
- Add 11.6 µl of the tag (6mM stock solution in DMSO) and vortex.
- Add 11.6 µl of fresh TCEP solution without vortexing.
- Add 35 µl of ligand solution and vortex.
- Add 11.6 µl of copper (II) sulfate (50 mM stock in water (12.5 mg/ml)) and vortex.
- Leave the tubes at room temperature for 1 h (vortex after 30 min). At this stage, the proteins will start to precipitate and the solution will turn cloudy.
- Centrifuge for 4 min at 6,500g and 4°C. A protein pellet will form.
- Remove the supernatant and add 500 µl of cold MeOH. Sonicate for 3–4 s. (Ensure that the MeOH used for washing the protein pellet is cooled on ice before use. The use of warm MeOH results in loss of protein from the pellet.)
- Rotate at 4°C for 10 min and centrifuge for 4 min at 6,500g and 4°C.
- Remove the supernatant and repeat the wash with 500 µl of cold MeOH (step 11 and 12).
- Remove the supernatant, add 1 ml of 1.2% SDS/PBS, sonicate for 3–4 s and heat to 80–90°C for 5 min.
- Transfer the solution to a 15 ml conical tube and dilute to 0.2% SDS with 5 ml of PBS.
- Samples can be stored at -80°C for several days.
For more details refer to the following reference:
- Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)-a general method for mapping sites of probe modification in proteomes; Eranthie Weerapana, Anna E. Speers and Benjamin F. Cravatt; NATURE PROTOCOLS, 2007; 2(6): 1414-1425; DOI:10.1038/nprot.2007.194
.