Welcome to Iris Biotech
For better service please confirm your country and language we detected.

For better service please confirm your country and language we detected.

Thank you very much for your interest in our products. All prices listed on our website are ex-works, Germany, and may attract customs duties when imported.
You may/will be contacted by the shipping company for additional documentation that may be required by the US Customs for clearance.
We offer you the convenience of buying through a local partner, Peptide Solutions LLC who can import the shipment as well as prepay the customs duties and brokerage on your behalf and provide the convenience of a domestic sale.
Continue to Iris Biotech GmbHSend request to US distributorPublished on 22.08.2023

Cell-free protein synthesis systems, also termed in vitro translation systems, expand the diversity of protein expression in a remarkable way. The intense development and analysis of pro- and eukaryotic in vitro translation systems are permanently growing fields of research. Open cell-free systems are designed to express complex membrane proteins, glycoproteins, and cytosolic proteins with the addition of posttranslational modifications onto the growing nascent polypeptide chain. The development of translationally active cell lysates into highly productive systems, the generation and optimization of particular DNA and RNA templates for different cell-free systems and the optimization of reaction conditions in newly developed in vitro translation systems are part of intense research investigations.
Typically, cell-free systems use crude cell extracts prepared from cells of choice by lysis followed by many steps of washing to remove the cell debris and genomic DNA. These cell extracts can be stored at − 80 °C for years and can be used by thawing just before the reaction. Such extracts contain all the principal components necessary for translation, such as aminoacyl-tRNA synthetase (AAS), ribosomes, and factors necessary for elongation, initiation, and transcription. Protein synthesis can be realized by combining cell extracts with necessary substrates like amino acids, energy substrates, DNA, cofactors, salts, and nucleotides. Depending on the biochemical properties of the protein and its end application, the appropriate cell-free system can be selected.

Cell-free reaction starts with fermentation of eukaryotic cells followed by their collection, disruption, and purification. The cell lysate then contains ribosomes, microsomes, aminoacyl-tRNA-synthetases and translation factors. This will be combined with the mRNA of the to be produced protein and completed with amino acids and energy supplements.
Cell-free protein synthesis is a fast protein production system since it does not require transfection or cell culture and lacks cell viability constraints. The cell-free system can be applied for the optimization of DNA constructs prior to in vivo expression, the production of difficult-to-express proteins such as toxic proteins and membrane-spanning proteins as well as the synthesis of proteins with improved properties by incorporating non-canonical amino acids.
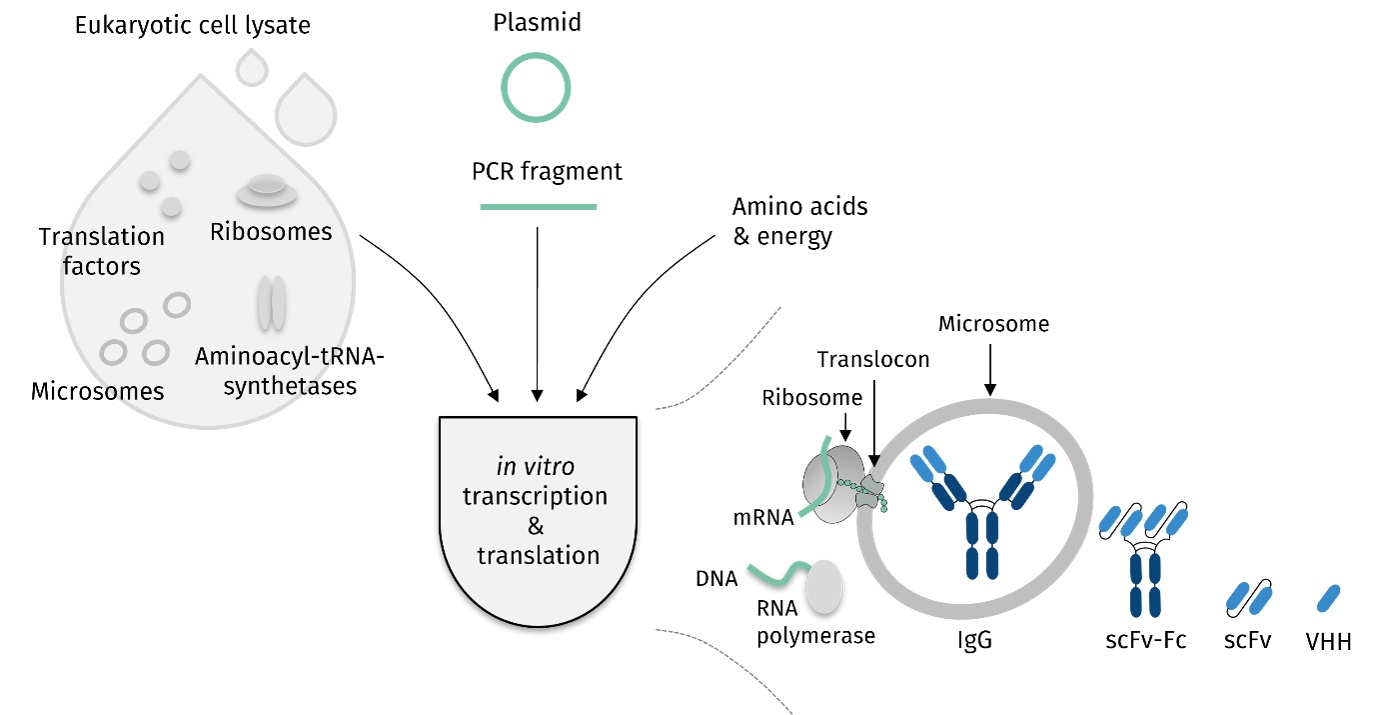
Cell-free antibody synthesis is typically performed in translationally active lysates from CHO cells. Various antibody formats, such as single-chain antibodies and nanobodies, can also be produced in cell-free systems. Starting from the plasmid or linear PCR template, various antibody formats can be produced cell-free in a high-throughput compatible manner.
Especially the last-mentioned incorporation of non-standard amino acids has gained increasing attention over the past years. As a result, improved methods are now available, enabling the efficient and directed co-translational incorporation of various non-standard amino acids to equip proteins with desired characteristics. In this context, the utilization of cell-free protein synthesis is particularly useful due to the direct accessibility of the translational machinery and synthesized proteins without having to maintain a vital cellular host.
A widely used example of such a non-canonical amino acid is p-azido-L-phenylalanine (HAA1850) (Phe(N3)), which can be used for protein interaction studies due to its photosensitivity. Furthermore, Phe(N3) is known to selectively react under physiological conditions via a set of different reactions such as Staudinger ligation, copper-catalyzed alkyne-azide cycloadditions and copper-independent strain-promoted alkyne-azide cycloadditions.
In this context, bioorthogonal chemistry facilitates functional analysis of proteins produced in cell-free systems.

Via addition of appropriate non-canonical amino acids, like p-azidophenylalanine (Phe(N3)) an orthogonal azido function can be implemented on any desired position within the protein, which serves as point of conjugation for subsequent reaction with linkers carrying various types of organic or inorganic cargo molecules.
Applications of non-canonical amino acid incorporation in cell-free systems include the development of novel therapeutics, polymers, and enzymes. In particular, the cell-free synthesis of homogenous antibody–drug conjugates is a promising tool to generate highly specific cancer therapeutics.
→ For more details on cell-free bioengineering and Linkerology®, click here!
→ Interested in linker technologies? Download our brochure Linkerology®
→ For any protein of your interest with/without Linkerology® please inquire for a custom synthesis!
References:
Evaluation of the Ion Channel Assembly in a Eukaryotic Cell-Free System Focusing on Two-Pore Domain Potassium Channels K2P; J. Ullrich, C. Ohlhoff, S. K. Dondapati, A. Zemella, S. Kubick; International Journal of Molecular Sciences 2023; 24: 6299. https://doi.org/10.3390/ijms24076299
Enriched cell-free and cell-based native membrane derived vesicles (nMV) enabling rapid in-vitro electrophysiological analysis of the voltage-gated sodium channel 1.5; Y. Pandey, S. K. Dondapati, S. Kubick; Biochim Biophys Acta Biomembr 2023; 1865: 184144. https://doi.org/10.1016/j.bbamem.2023.184144
Rapid One-Step Capturing of Native, Cell-Free Synthesized and Membrane-Embedded GLP-1R; L. Haueis, M. Stech, E. Schneider, T. Lanz, N. Hebel, A. Zemella, S. Kubick; Int J Mol Sci 2023; 24: 2808. https://doi.org/10.3390/ijms24032808
Cell-Free Systems Enable the Production of AB(5) Toxins for Diagnostic Applications; F. Ramm, L. Jack, D. Kaser, J. L. Schlosshauer, A. Zemella, S. Kubick; Toxins (Basel) 2022; 14: 233. https://doi.org/10.3390/toxins14040233
The Potential of Eukaryotic Cell-Free Systems as a Rapid Response to Novel Zoonotic Pathogens: Analysis of SARS-CoV-2 Viral Proteins; F. Ramm, S. K. Dondapati, H. A. Trinh, D. Wenzel, R. M. Walter, A. Zemella, S. Kubick; Front Bioeng Biotechnol 2022; 10: 896751. https://doi.org/10.3389/fbioe.2022.896751
Synthesis of an Anti-CD7 Recombinant Immunotoxin Based on PE24 in CHO and E. coli Cell-Free Systems; S. K. Krebs, M. Stech, F. Jorde, N. Rakotoarinoro, F. Ramm, S. Marinoff, S. Bahrke, A. Danielczyk, D. A. Wustenhagen, S. Kubick; Int J Mol Sci 2022; 23: 13697. https://doi.org/10.3390/ijms232213697
A Cell-free Expression Pipeline for the Generation and Functional Characterization of Nanobodies; L. Haueis, M. Stech, S. Kubick; Front Bioeng Biotechnol 2022; 10: 896763. https://doi.org/10.3389/fbioe.2022.896763
Synthesis of Fluorescently Labeled Antibodies Using Non-Canonical Amino Acids in Eukaryotic Cell-Free Systems; M. Stech, N. Rakotoarinoro, T. Teichmann, A. Zemella, L. Thoring, S. Kubick; Structural Proteomics: High-Throughput Methods R. J. Owens 2021: 175-190. https://doi.org/10.1007/978-1-0716-1406-8_9
Cell-free production of pore forming toxins: Functional analysis of thermostable direct hemolysin from Vibrio parahaemolyticus; S. K. Dondapati, D. A. Wustenhagen, E. Strauch, S. Kubick; Eng Life Sci 2018; 18: 140-148. https://doi.org/10.1002/elsc.201600259
Cell-free synthesis of functional antibodies using a coupled in vitro transcription-translation system based on CHO cell lysates; M. Stech, O. Nikolaeva, L. Thoring, W. F. M. Stocklein, D. A. Wustenhagen, M. Hust, S. Dubel, S. Kubick; Sci Rep 2017; 7: 12030. https://doi.org/10.1038/s41598-017-12364-w
Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems; A. Zemella, L. Thoring, C. Hoffmeister, S. Kubick; Chembiochem : a European journal of chemical biology 2015; 16: 2420-31. https://doi.org/10.1002/cbic.201500340
Cell-free eukaryotic systems for the production, engineering, and modification of scFv antibody fragments; M. Stech, M. Hust, C. Schulze, S. Dübel, S. Kubick; Eng Life Sci 2014; 14: 387-398. https://doi.org/10.1002/elsc.201400036